
Abstract:
Genetically modified organisms are produced by modifying or altering their genetic material by means of Genetic Engineering technique and which does not occur naturally.
DNA based identification is more superior and widely accepted compared to protein based approach because of their limitations.
In the DNA based method, DNA is extracted from homogeneous test material. The quantity and quality of the extracted DNA are evaluated for the testing the suitability of PCR amplification.
Then a series of (PCR) tests performed for screening of the Targets specific genetic elements and constructs that are most frequently present in commercialized GM crops.
The (combined) outcome of these tests then systematically compared with a predicted matrix comprising of a list of possible genetic construct or element with the respective GM event.
Key word: GM (genetically modified), DNA (Deoxyribonucleic acid), Probe, Ct value, RT- PCR (real time PCR).
Introduction:
Genetically modified organisms (GMO) were introduced into the food chain over 20 years ago with the intention to provide special beneficial outcome like more productivity, nutritional value, insect resistance, extended shelf life etc. Since the use of genetically modified microorganisms is somewhat controversial. Though the cultivation of GM crops is concentrated in certain regions (e.g. U.S., Argentina, Brazil), they are spread globally by trading, transportation and storage either intentionally or inadvertently by contamination of genetically modified organism (GMO) free goods.
In GMO one or more targeted genetic elements transfer from one organism to another organism that may not related to each other. As for example, a bacteria gene to plant or animal genome, Viral gene to plant or animal genome, Bacteria gene to another bacterial genome.
To insert and activate respective genetic material, a construct of the gene(s) of interest is combined with a promoter and terminator. Sometimes marker sequences are also designed and transformed into plant cells. Once introduced, they stabilize and integrate functionally in the plant genome.
How to detect GMO?
Since the difference between the native plant and GM counterpart is based on DNA, the combination of plant or host genome and integrated construct enables different levels of specificity that can be used for detection in three of the following ways: Element specific, Construct specific, Event specific as illustrated in below figure.
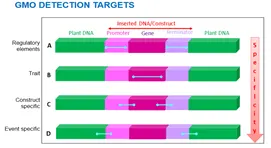
As per the information available in International Service for the Acquisition of Agri-biotech Applications database (ISAAA), there are more than 505 event among 25 different crops worldwide and it is not possible to identify all the event. So most acceptable approach is to target the most frequently available genetic elements such as:
Material & method:
Test sample: Soya with at 0.1% GM content (having TNOS & P35S target element) & Sample with 0% GM content as a negative sample.
Equipment & reagents:
Balance, mixer grinder, centrifuge, vortex, Laminar flow, UV spectrophotometer, Real time PCR, Autoclave.
DNA extraction kit, PCR master mix, PCR primer, PCR probe.
DNA Extraction:
DNA extraction is done after cell lysis with buffer, followed by repeated washing and centrifuge by using Column based extraction kit.
Estimating the quantity & quality of extracted DNA for using with PCR:
DNA quality & quantity is estimated by using following factors-
Take the absorbance at 260 nm & 280 nm by UV-Vis spectrophometer
DNA Quantity (µg/ml) = A260 * 50 (as A260 = 1 = 50 µg/ml).
Pure DNA has an A260/A280 ratio of 1.7–1.9.
PCR reaction set up:
Working stock of specific primer & probe are prepared by diluting at a particular level for the respective target genetic element.
Extracted DNA is also diluted to get the required amount of DNA in the final reaction to achieve the desired LOD (0.1%).
For every cycle, each sample is run at least in duplicate along with the below controls:
Negative control: DNA isolated from the certified reference material containing no target element.
Positive control: DNA isolated from the certified reference material containing the target element at a level of LOD.
Extraction Control: DNA isolate containing reference gene from the tested matrix such as Lectin gene (Le1) for Soya, Alcohol dehydrogenase 1 (adh1) for Maize.
Positive control: IRMM certified material ERM-BF410cp (Level 1, 0.1% GMO) for P35S & TNOS
Target specific primer & probe selection is done based on the information available in online database.
Data interpretation :
The presence of target element is determined based on the PCR amplification signal in the target channel (Green channel for P35 target element & Le1; Yellow channel for TNOS) & corresponding threshold cycle (Ct).
Good amplification curve with a Ct value up to 34 of target along with the acceptable results of controls indicate the presence of the target element or presence of GM element.
Conclusion:
Present study produced good amplification curve with acceptable Ct value with 33.4 in both target channels (FAM or Green for P35S & HEX or Yellow for TNOS) & there is no amplification observed for blank sample where as extraction control (Le1 gene) provide good amplification with Ct a value of 20.78.
So it can be concluded that the method is suitable for the identify the P35S & TNOS element from any soya sample.
The same approach shall be applied for the detection of other elements with the use of specific combination of primers & probe. More PCR analysis should be done if identification of particular event is desired.
Prepared By: Sukanta Palui & Ipsita Biswas (Udayan Lab, Microbiology Division)

Chief Operation, FAMD, Tata Steel Limited..

Sr. General Manager,, Emirates Trading Agency L.L.C..

Mines Manager, Hindustan Zinc Limited, a Vedanta Company.

General Manager, Stevin Rock L.L.C..

Executive Vice President (Works),, DCW Limited.

AVP – Coal Quality & Sales Compliance Head,, PT Indo Tambangraya Megah Tbk (BANPU).

Laboratory Head, MMX.

Shipping Administrator, Mount Gibson Iron Limited.

Senior Director – Asia Pacific Iron Ore Sales,, Cliffs Natural Resources Pty Ltd..

Member, Compass Group (India) Pvt. Ltd.

Posted on January 16 2026 By Mitra S.K ADMIN
Read More
Posted on January 16 2026 By Mitra S.K ADMIN

Posted on January 16 2026 By Mitra S.K ADMIN

Posted on January 16 2026 By Mitra S.K ADMIN

Posted on January 16 2026 By Mitra S.K ADMIN

Posted on December 16 2025 By Mitra S.K ADMIN

Posted on December 16 2025 By Mitra S.K ADMIN
Posted on December 16 2025 By Mitra S.K ADMIN

Posted on December 11 2025 By Mitra S.K ADMIN
![Estimating Cobalt by UV-Vis Spectroscopy: The [CoCl?]²? Acetone Method](https://mitrask.com/uploads/blogs/1764834098Estimating%20Cobalt.png)
Posted on December 04 2025 By Mitra S.K ADMIN
Posted on December 04 2025 By Mitra S.K ADMIN

Posted on November 12 2025 By Mitra S.K ADMIN

Posted on September 23 2025 By Mitra S.K ADMIN

Posted on August 01 2025 By Mitra S.K ADMIN

Posted on July 25 2025 By Mitra S.K ADMIN

Posted on July 18 2025 By Mitra S.K ADMIN

Posted on July 01 2025 By Mitra S.K ADMIN

Posted on May 22 2025 By Mitra S.K ADMIN

Posted on January 24 2025 By Mitra S.K ADMIN

Posted on January 24 2025 By Mitra S.K ADMIN

Posted on December 31 2024 By Mitra S.K ADMIN

Posted on December 31 2024 By Mitra S.K ADMIN
Posted on December 31 2024 By Mitra S.K ADMIN

Posted on December 31 2024 By Mitra S.K ADMIN

Posted on December 31 2024 By Mitra S.K ADMIN

Posted on December 03 2024 By Mitra S.K ADMIN

Posted on October 17 2024 By Mitra S.K ADMIN

Posted on October 04 2024 By Mitra S.K ADMIN

Posted on September 13 2024 By Mitra S.K ADMIN

Posted on August 27 2024 By Mitra S.K ADMIN

Posted on August 23 2024 By Mitra S.K ADMIN

Posted on June 27 2024 By Mitra S.K ADMIN

Posted on June 22 2024 By Mitra S.K ADMIN

Posted on June 15 2024 By Mitra S.K ADMIN

Posted on May 24 2024 By Mitra S.K ADMIN
Posted on May 17 2024 By Mitra S.K ADMIN

Posted on May 09 2024 By Mitra S.K ADMIN

Posted on April 20 2024 By Mitra S.K ADMIN

Posted on April 13 2024 By Mitra S.K ADMIN

Posted on April 30 2024 By Mitra S.K ADMIN

Posted on April 29 2024 By Mitra S.K ADMIN

Posted on December 30 2023 By Mitra S.K ADMIN

Posted on December 30 2023 By Mitra S.K ADMIN

Posted on December 30 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 27 2023 By Mitra S.K ADMIN

Posted on December 26 2023 By Mitra S.K ADMIN

Posted on April 05 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on November 28 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on April 06 2022 By Mitra S.K ADMIN

Posted on November 28 2022 By Mitra S.K ADMIN

Posted on June 14 2022 By Mitra S.K ADMIN






